2 Overview
This part of the book teaches you how to leverage the plotly R package to create a variety of interactive graphics. There are two main ways to creating a plotly object: either by transforming a ggplot2 object (via ggplotly()) into a plotly object or by directly initializing a plotly object with plot_ly()/plot_geo()/plot_mapbox(). Both approaches have somewhat complementary strengths and weaknesses, so it can pay off to learn both approaches. Moreover, both approaches are an implementation of the Grammar of Graphics and both are powered by the JavaScript graphing library plotly.js, so many of the same concepts and tools that you learn for one interface can be reused in the other.
The subsequent chapters within this ‘Creating views’ part dive into specific examples and use cases, but this introductory chapter outlines some over-arching concepts related to plotly in general. It also provides definitions for terminology used throughout the book and introduces some concepts useful for understanding the infrastructure behind any plotly object. Most of these details aren’t necessarily required to get started with plotly, but it will envitably help you get ‘un-stuck’, write better code, and do more advanced things with plotly.
2.1 Intro to plot_ly()
Any graph made with the plotly R package is powered by the JavaScript library plotly.js. The plot_ly() function provides a ‘direct’ interface to plotly.js with some additional abstractions to help reduce typing. These abstractions, inspired by the Grammar of Graphics and ggplot2, make it much faster to iterate from one graphic to another, making it easier to discover interesting features in the data (Wilkinson 2005); (Wickham 2009). To demonstrate, we’ll use plot_ly() to explore the diamonds dataset from ggplot2 and learn a bit how plotly and plotly.js work along the way.
# load the plotly R package
library(plotly)
# load the diamonds dataset from the ggplot2 package
data(diamonds, package = "ggplot2")
diamonds
#> # A tibble: 53,940 x 10
#> carat cut color clarity depth table price x
#> <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl>
#> 1 0.23 Ideal E SI2 61.5 55 326 3.95
#> 2 0.21 Prem… E SI1 59.8 61 326 3.89
#> 3 0.23 Good E VS1 56.9 65 327 4.05
#> 4 0.290 Prem… I VS2 62.4 58 334 4.2
#> 5 0.31 Good J SI2 63.3 58 335 4.34
#> 6 0.24 Very… J VVS2 62.8 57 336 3.94
#> # … with 5.393e+04 more rows, and 2 more variables:
#> # y <dbl>, z <dbl>If we assign variable names (e.g., cut, clarity, etc) to visual properties (e.g., x, y, color, etc) within plot_ly(), as done in Figure 2.1, it tries to find a sensible geometric representation of that information for us. Shortly we’ll cover how to specify these geometric representations (as well as other visual encodings) to create different kinds of charts.
# create three visualizations of the diamonds dataset
plot_ly(diamonds, x = ~cut)
plot_ly(diamonds, x = ~cut, y = ~clarity)
plot_ly(diamonds, x = ~cut, color = ~clarity, colors = "Accent")
FIGURE 2.1: Three examples of visualizing categorical data with plot_ly(): (top) mapping cut to x yields a bar chart, (middle) mapping cut & clarity to x & y yields a heatmap, and (c) mapping cut & clarity to x & color yields a dodged bar chart.
The plot_ly() function has numerous arguments that are unique to the R package (e.g., color, stroke, span, symbol, linetype, etc) and make it easier to encode data as visual properties. By default, these arguments map values to a visual range defined by the plural form of the argument (e.g., colors, strokes, spans, symbols, linetypes, etc). Figure 2.2 depicts how the combination of color (e.g., clarity levels) and colors (e.g., the "Accent" color palette from the RColorBrewer package) can be used to specify a mapping from data values to color codes. Besides providing a palette by name, one may also provide their own mapping function (e.g., colorRamp()), or even a set of custom color codes. See Chapter 3 for many more examples and Chapter 27 for more details about working with colors.
FIGURE 2.2: Mapping data values to a visual color range.
Since these arguments, by default, map values to a visual range, you will obtain unexpected results if you try to specify the visual range directly, as in the top portion of Figure 2.3. If you want to specify the visual range directly, use the I() function to declare this value to be taken ‘AsIs’, as in the bottom portion of Figure 2.3. As discussed in more detail in section 2.2, these ‘aesthetic mapping’ arguments are not officially part of plotly.js, but the R package transform this information into valid plotly.js figures. Many other examples in the book will leverage these arguments, especially Chapter 3. Another resource to learn more details about these arguments (especially their defaults) is the R documentation page available by entering help(plot_ly) in your R console.
# doesn't produce black bars
plot_ly(diamonds, x = ~cut, color = "black")
# produces red bars with black outline
plot_ly(diamonds, x = ~cut, color = I("red"), stroke = I("black"), span = I(2))
FIGURE 2.3: Using I() to supply visual properties directly instead of mapping values to a visual range. In the top portion of this figure, the value 'black' is being mapped to a visual range spanned by colors (which, for discrete data, defaults to 'Set2').
The plotly package takes a purely functional approach to a layered grammar of graphics (Wickham 2010).2 The purely functional part means, (almost) every function anticipates a plotly object as input to it’s first argument and returns a modified version of that plotly object. Furthermore, that modification is completely determined by the input values to the function (i.e., it doesn’t rely on any side-effects, unlike, for example, base R graphics). For a quick example, the layout() function anticipates a plotly object in it’s first argument and it’s other arguments add and/or modify various layout components of that object (e.g., the title):
layout(
plot_ly(diamonds, x = ~cut),
title = "My beatiful histogram"
)For more complex plots that modify a plotly graph many times over, code written in this way can become cumbersome to read. In particular, we have to search for the inner-most part of the R expression, then work outwards towards the end result. The %>% operator from the magrittr package allows us to re-arrange this code so that we can read the sequence of modifications from left-to-right rather than inside-out (Bache and Wickham 2014). The %>% operator enable this by placing the object on the left-hand side of the %>% into the first argument of the function of the right-hand side.
diamonds %>%
plot_ly(x = ~cut) %>%
layout(title = "My beatiful histogram")In addition to layout() for adding/modifying part(s) of the graphs’s layout, there are also a family of add_*() functions (e.g., add_histogram(), add_lines(), etc) that define how to render data into geometric objects. Borrowing terminology from the layered grammar of graphics, these functions add a graphical layer to a plot. A layer can be thought of as a group of graphical elements that can be sufficiently described using only 5 components: data, aethestic mappings (e.g., assigning clarity to color), a geometric representation (e.g. rectangles, circles, etc), statistical transformations (e.g., sum, mean, etc), and positional adjustments (e.g., dodge, stack, etc). If you’re paying attention, you’ll notice that in the examples thus far, we have not specified a layer! The layer has been added for us automatically by plot_ly(). To be explicit about what plot_ly(diamonds, x = ~cut) generates, we should add a add_histogram() layer:
diamonds %>%
plot_ly() %>%
add_histogram(x = ~cut)As you’ll learn more about in Section 5, plotly has both add_histogram() and add_bars(). The difference is that add_histogram() performs statistics (i.e., a binning algorithm) dynamically in the web browser, whereas add_bars() requires the bar heights to be pre-specified. That means, to replicate the last example with add_bars(), the number of observations must be computed ahead-of-time.
diamonds %>%
dplyr::count(cut) %>%
plot_ly() %>%
add_bars(x = ~cut, y = ~n)There are numerous other add_*() functions that calculate statistics in the browser (e.g., add_histogram2d(), add_contour(), add_boxplot(), etc), but most other functions aren’t considered statistical. Making the distinction might not seem useful now, but they have their own respective trade-offs when it comes to speed and interactivity. Generally speaking, non-statistical layers will be faster and more responsive at run-time (since they require less computational work), whereas the statistical layers allow for more flexibility when it comes to client-side interactivity, as covered in section 16. Practically speaking, the difference in performance is often negligible – the more common bottleneck occurs when attempting to render lots of graphical elements at a time (e.g., a scatterplot with a million points). In those scenarios, you likely want to render your plot in Canvas rather than SVG (the default) via toWebGL() – for more information on improving performance, see 24.
In many scenarios, it can be useful to combine multiple graphical layers into a single plot. In this case, it becomes useful to know a few things about plot_ly():
- Arguments specified in
plot_ly()are global, meaning that any downstreamadd_*()functions inherit these arguments (unlessinherit = FALSE). - Data manipulation verbs from the dplyr package may be used to transform the
dataunderlying a plotly object.3
Using these two properties of plot_ly(), Figure 2.4 demonstrates how we could leverage these properties of plot_ly() to do the following:
- Globally assign
cuttox. - Add a histogram layer (inherits the
xfromplot_ly()). - Use dplyr verbs to modify the
dataunderlying the plotly object. Here we just count the number of diamonds in eachcutcategory. - Add a layer of text using the summarized counts. Note that the global
xmapping, as well as the other mappings local to this text layer (textandy), reflect data values from step 3.
library(dplyr)
diamonds %>%
plot_ly(x = ~cut) %>%
add_histogram() %>%
group_by(cut) %>%
summarise(n = n()) %>%
add_text(
text = ~scales::comma(n), y = ~n,
textposition = "top middle",
cliponaxis = FALSE
)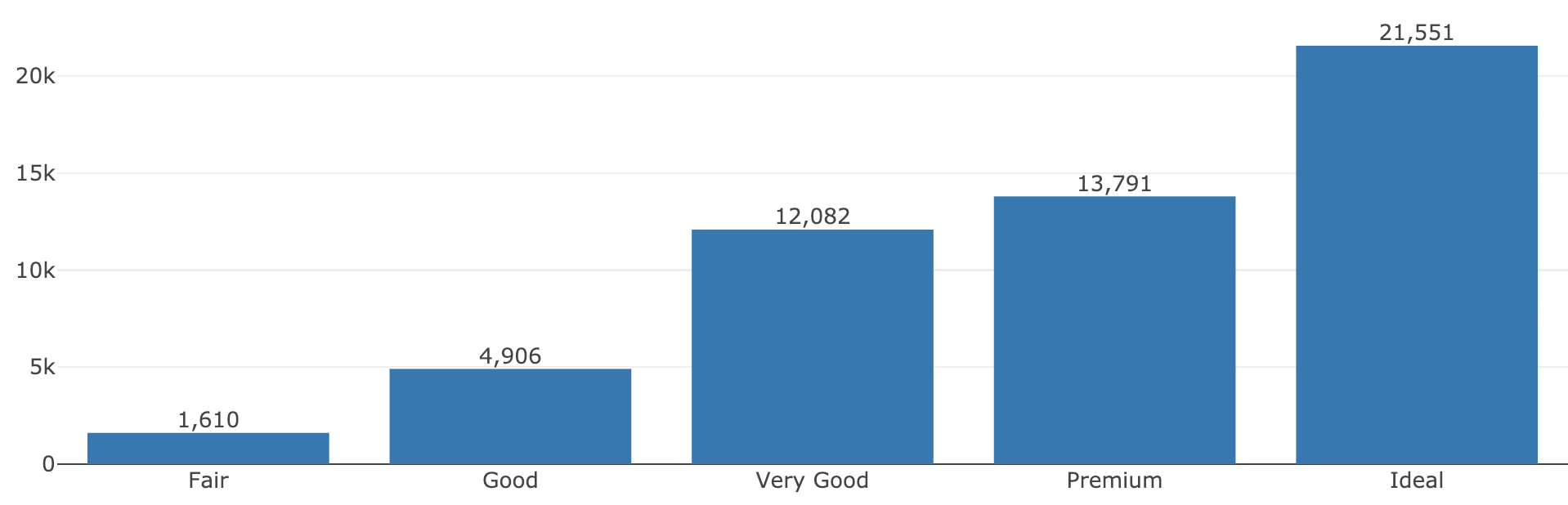
FIGURE 2.4: Using add_histogram(), add_text(), and dplyr verbs to compose a plot that leverages a raw form of the data (e.g., histogram) as well as a summarized version (e.g., text labels).
Before using multiple add_*() in a single plot, make sure that you actually want to show those layers of information on the same set of axes. If it makes sense to display the information on the same axes, consider making multiple plotly objects and combining them into as grid-like layout using subplot(), as described in section 13. Also, when using dplyr verbs to modify the data underlying the plotly object, you can use the plotly_data() function to obtain the data at any point in time. This can be helpful for finding why something has
diamonds %>%
plot_ly(x = ~cut) %>%
add_histogram() %>%
group_by(cut) %>%
summarise(n = n()) %>%
plotly_data()
#> # A tibble: 5 x 2
#> cut n
#> <ord> <int>
#> 1 Fair 1610
#> 2 Good 4906
#> 3 Very Good 12082
#> 4 Premium 13791
#> 5 Ideal 21551This introduction to plot_ly() has mainly focused on concepts unique to the R package plotly that are generally useful for creating most kinds of data views. The section outlines how plotly generates plotly.js figures and how to inspect the underlying data structure that plotly.js uses to render the graph. Not only is this information useful for debugging, but it’s also a nice way to learn how to work with plotly.js directly, which you may need to improve performance in shiny apps (Chapter 17.3.1) and/or for adding custom behavior with JavaScript (Chapter 18).
2.2 Intro to plotly.js
To recreate the plots in Figure 2.1 using plotly.js directly, it would take significantly more code and knowledge of plotly.js. That being said, learning how plotly generates the underlying plotly.js figure is a useful introduction to plotly.js itself, and knowledge of plotly.js becomes useful when you need more flexible control over plotly. As Figure 2.5 illustrates, when you print any plotly object, the plotly_build() function is applied to that object, and that generates an R list which adheres to a syntax that plotly.js understands. This syntax is a JavaScript Object Notation (JSON) specification that plotly.js uses to represent, seralize, and render web graphics. A lot of documentation you’ll find online about plotly (e.g., the online figure reference) implictly refers to this JSON specification, so it can helpful to know how to “work backwards” from that documentation (i.e., translate JSON into to R code). If you’d like to learn details about mapping between R and JSON, section 19 provides an introduction aimed at R programmers, and Ooms (2014) provides a cohesive overview of the jsonlite package, which is what plotly uses to map between R and JSON.
FIGURE 2.5: A diagram of what happens when you print a plotly graph.
For illustration purposes, Figure 2.5 shows how this workflow applies to a simple bar graph (with values directly supplied instead of a data column name reference like Figure 2.1), but the same concept applies for any graph created via plotly. As the diagram suggests, both the plotly_build() and plotly_json() functions can be used to inspect the underlying data structure on both the R and JSON side of things. For example, Figure 2.6 shows the data portion of the JSON created for the last graph in Figure 2.6.
p <- plot_ly(diamonds, x = ~cut, color = ~clarity, colors = "Accent")
plotly_json(p)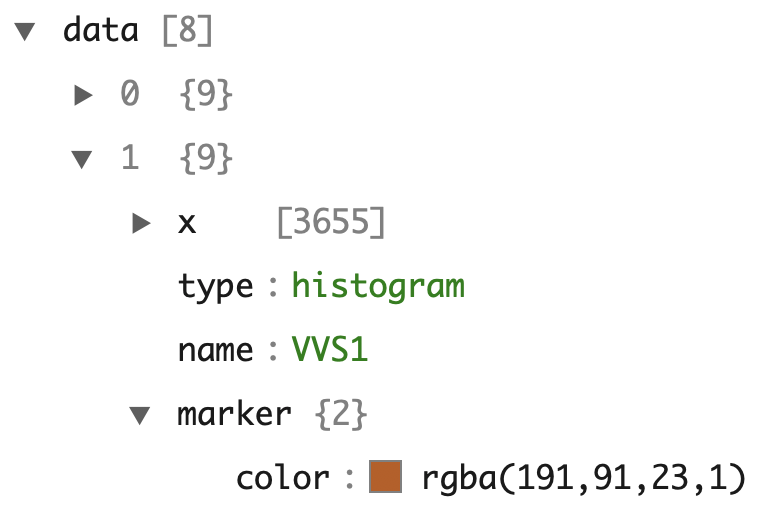
FIGURE 2.6: A portion of the JSON data behind the bottom plot of Figure 2.1. This dodged bar chart has 8 layers of data (i.e., 8 traces) – one for each level of clarity.
In plotly.js terminology, a figure has two key components: data (aka, traces) and a layout. A trace defines a mapping from data and visuals.4 Every trace has a type (e.g., histogram, pie, scatter, etc) and the trace type determines what other attributes (i.e., visual and/or interactive properties, like x, hoverinfo, name) are available to control the trace mapping. That is, not every trace attribute is available to every trace type, but many attributes (e.g., the name of the trace) are available in every trace type and serve a similar purpose. From Figure 2.6 we can see that it takes multiple traces to generate the dodged bar chart, but instead of clicking through JSON viewer, sometimes it’s easier to use plotly_build() and compute on the plotly.js figure definition to verify certain things exist. Since plotly uses the htmlwidgets standard5, the actual plotly.js figure definition appears under a list element named x (Vaidyanathan et al. 2016).
# use plotly_build() to get at the plotly.js definition
# behind *any* plotly object
b <- plotly_build(p)
# Confirm there 8 traces
length(b$x$data)
#> [1] 8
# Extract the `name` of each trace. plotly.js uses `name` to
# populate legend entries and tooltips
purrr::map_chr(b$x$data, "name")
#> [1] "IF" "VVS1" "VVS2" "VS1" "VS2" "SI1" "SI2" "I1"
# Every trace has a type of histogram
unique(purrr::map_chr(b$x$data, "type"))
#> [1] "histogram"Here we’ve learned that plotly creates 8 histogram traces to generate the dodged bar chart: one trace for each level of clarity.6 Why one trace per category? As illustrated in Figure 2.7 there are two main reasons: to populate a tooltip and legend entry for each level of clarity level.
FIGURE 2.7: Leveraging two interactive features that require one trace per level of clarity: (1) Using ‘Compare data on hover’ mode to get counts for every level of clarity for a given level of cut and (2) Using the ability to hide/show clarity levels via their legend entries.
If we investigated further, we’d notice that color and colors are not officially part of the plotly.js figure definition – the plotly_build() function has effectively transformed that information into a sensible plotly.js figure definition (e.g., marker.color contains the actual bar color codes). In fact, the color argument in plot_ly() is just one example of an abstraction the R package has built on top of plotly.js to make it easier to map data values to visual attributes, and many of these are covered in Section 3.
2.3 Intro to ggplotly()
The ggplotly() function from the plotly package has the ability to translate ggplot2 to plotly. This functionality can be really helpful for quickly adding interactivity to your existing ggplot2 workflow.7 Moreover, even if you know plot_ly() and plotly.js well, ggplotly() can still be desirable for creating visualizations that aren’t necessarily straight-forward to acheive without it. To demonstrate, let’s explore the relationship between price and other variables from the well-known diamonds dataset.
Hexagonal binning (i.e., geom_hex()) is useful way to visualize a 2D density8, like the relationship between price and carat as shown in Figure 2.8. From Figure 2.8, we can see there is a strong positive linear relationship between the log of carat and price. It also shows that many the carat is only rounded to a particular number (indicated by the light blue bands) and no diamonds are priced around $1500. Making this plot interactive makes it easier to decode the hexagonal colors into the counts that they represent.
p <- ggplot(diamonds, aes(x = log(carat), y = log(price))) +
geom_hex(bins = 100)
ggplotly(p)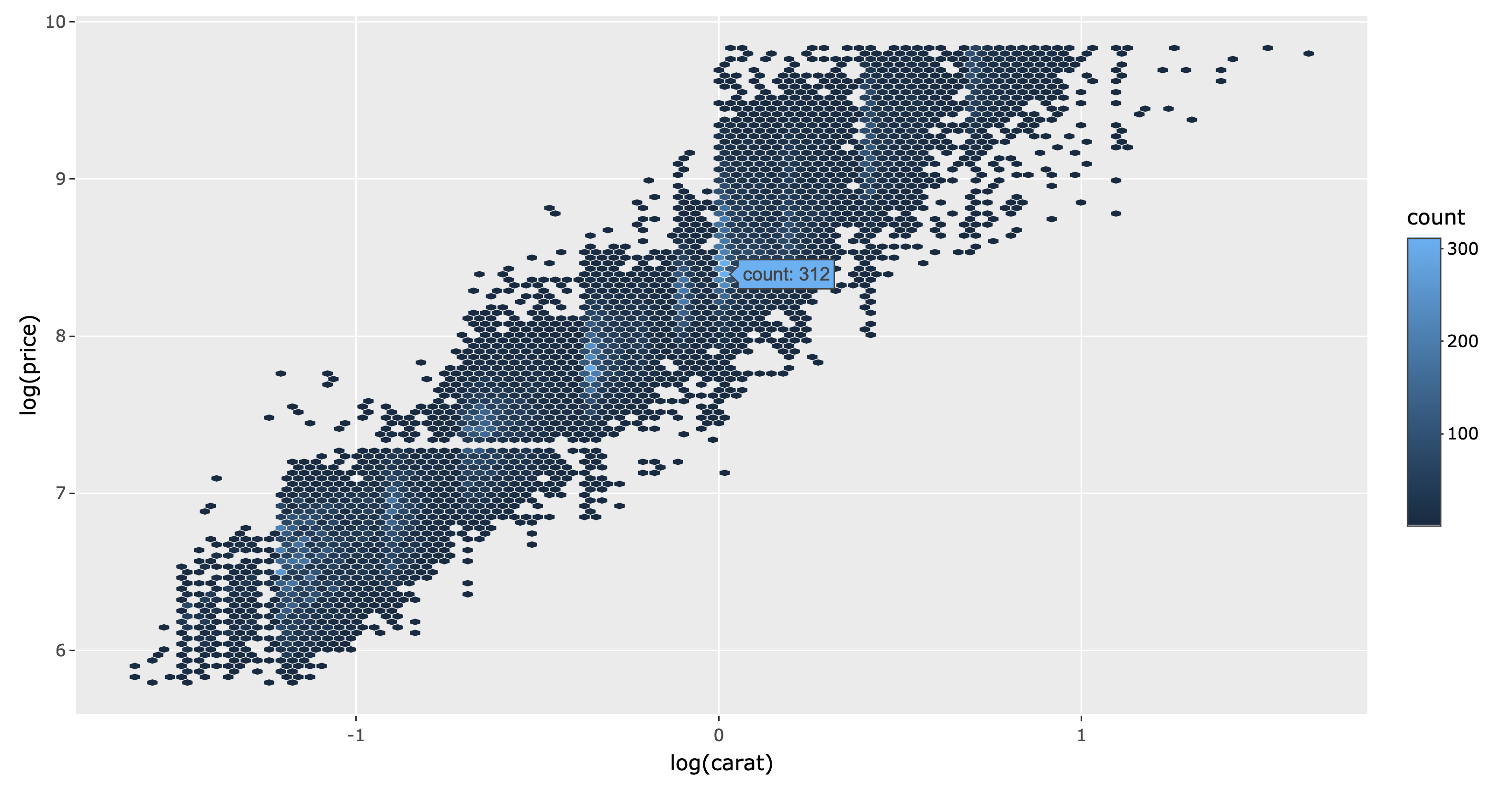
FIGURE 2.8: A hexbin plot of diamond carat versus price.
I often use ggplotly() over plot_ly() to leverage ggplot2’s consistent and expressive interface for exploring statistical summaries across groups. For example, by including a discrete color variable (e.g., cut) with geom_freqpoly(), you get a frequency polygon for each level of that variable. This ability to quickly generate visual encodings of statisitical summaries across an arbitrary number of groups works for basically any geom (e.g. geom_boxplot(), geom_histogram(), geom_density(), etc) and is a key feature of ggplot2.
p <- ggplot(diamonds, aes(x = log(price), color = clarity)) +
geom_freqpoly()
ggplotly(p)FIGURE 2.9: Frequency polygons of diamond price by diamond clarity. This visualization indicates there may be significant main effects.
Now, to see how price varies with both cut and clarity, we could repeat this same visualization for each level of cut. This is where ggplot2’s facet_wrap() comes in handy. Moreover, to facilitate comparisons, we can have geom_freqpoly() display relative rather than absolute frequencies. By making this plot interactive, we can more easily compare particular levels of clarity (as shown in Figure 2.10) by leveraging the legend filtering capabilites.
p <- ggplot(diamonds, aes(x = log(price), color = clarity)) +
geom_freqpoly(stat = "density") +
facet_wrap(~cut)
ggplotly(p)FIGURE 2.10: Diamond price by clarity and cut.
In addition to supporting most of the ‘core’ ggplot2 API, ggplotly() can automatically convert any ggplot2 extension packages that return a ‘standard’ ggplot2 object. By standard, I mean that the object is comprised of ‘core’ ggplot2 data structures and not the result of custom geoms.9 Some great examples of R packages that extend ggplot2 using core data structures are ggforce, naniar, and GGally (Pedersen 2019); (Tierney et al. 2018); (Schloerke et al. 2016).
Figure 2.11 demonstrates another way of visualizing the same information found in Figure 2.10 using geom_sina() from the ggforce package (instead of geom_freqpoly()). This visualization jitters the raw data within the density for each group – allowing us not only only see where the majority observations fall within a group, but also across all across all groups. By making this layer interactive, we can query individual points for more information and zoom into interesting regions. The second layer of Figure 2.11 uses ggplot2’s stat_summary() to overlay a 95% confidence interval esimated via a bootstrap algorithm via the Hmisc package (Harrell Jr, Charles Dupont, and others. 2019).
p <- ggplot(diamonds, aes(x = clarity, y = log(price), color = clarity)) +
ggforce::geom_sina(alpha = 0.1) +
stat_summary(fun.data = "mean_cl_boot", color = "black") +
facet_wrap(~cut)
# WebGL is a lot more efficient at rendering lots of points
toWebGL(ggplotly(p))FIGURE 2.11: A sina plot of diamond price by clarity and cut.
As noted by Wickham and Grolemund (2018), it’s surprising that the diamond price would decline with an increase of diamond clarity. As it turns out, if we account for the carat of the diamond, then see that better diamond clarity does indeed lead to a higher diamond price, as shown in Figure 2.12. Seeing such a strong pattern in the residuals of simple linear model of carat vs price indicates that our model could be greatly improved by adding clarity as a predictor of price.
m <- lm(log(price) ~ log(carat), data = diamonds)
diamonds <- modelr::add_residuals(diamonds, m)
p <- ggplot(diamonds, aes(x = clarity, y = resid, color = clarity)) +
ggforce::geom_sina(alpha = 0.1) +
stat_summary(fun.data = "mean_cl_boot", color = "black") +
facet_wrap(~cut)
toWebGL(ggplotly(p))FIGURE 2.12: A sina plot of diamond price by clarity and cut, after accounting for carat.
As discussed in Section 16.4.7 the GGally package provides a convenient interface for making similar types of model diagnostic visualizations via the ggnostic() function. It also provides a convenience function for visualizating the coefficient estimates and their standard errors via the ggcoef() function. Figure 2.13 shows how injecting interactivity to this plot allows us to query exact values and zoom in on the most intere
library(GGally)
m <- lm(log(price) ~ log(carat) + cut, data = diamonds)
gg <- ggcoef(m)
# dynamicTicks means generate new axis ticks on zoom
ggplotly(gg, dynamicTicks = TRUE)
FIGURE 2.13: Zooming in on a coefficient plot generated from the ggcoef() function from the GGally package.
Although the diamonds dataset does not contain any missing values, it’s a very common problem in real data analysis problems. The naniar package provides a suite of computational and visual resources for working with and revealing structure in missing values. All the ggplot2 based visualizations return an object that can be converted by ggplotly(). Moreover, naniar provides a custom geom, geom_miss_point(), that can be useful for visualizing missingness structure. Figure 2.14 demonstrates this by introducing fake missing values to the diamond price.
library(naniar)
# fake some missing data
diamonds$price_miss <- ifelse(diamonds$depth > 60, diamonds$price, NA)
p <- ggplot(diamonds, aes(x = clarity, y = log(price_miss))) +
geom_miss_point(alpha = 0.1) +
stat_summary(fun.data = "mean_cl_boot", colour = "black") +
facet_wrap(~cut)
toWebGL(ggplotly(p))
FIGURE 2.14: Using the geom_miss_point() function from the naniar package to visualize missing values in relation to non-missing values. Missing values are shown in red.
In short, the ggplot2 ecosystem provides a world-class exploratory visualization toolkit, and having the ability to quickly insert interactivity such as hover, zoom, and filter via ggplotly() makes it even more powerful for exploratory analysis. In this introduction to ggplotly(), we’ve only seen relatively simple techniques that come for free out-of-the-box, but the true power of interactive graphics lies in linking multiple views. In that part of the book, you can find lots of examples of linking multiple (ggplotly() & plot_ly()) graphs purely client-side as well as with shiny.
It’s also worth mentioning that ggplotly() conversions are not always perfect and ggplot2 doesn’t provide an API for interactive features, so sometimes it’s desirable to modify the return values of ggplotly(). Section 33 talks generally about modifying the data structure underlying ggplotly() (which, by the way, uses the same a plotly.js figure definition as discussed in Section 2.2). Moreover, Section 25.2 outlines various ways to customize the tooltip that ggplotly() produces.
References
Wilkinson, Leland. 2005. The Grammar of Graphics (Statistics and Computing). Secaucus, NJ, USA: Springer-Verlag New York, Inc.
Wickham, Hadley. 2009. Ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. http://ggplot2.org.
Wickham, Hadley. 2010. “A Layered Grammar of Graphics.” Journal of Computational and Graphical Statistics 19 (1): 3–28.
Bache, Stefan Milton, and Hadley Wickham. 2014. Magrittr: A Forward-Pipe Operator for R. https://CRAN.R-project.org/package=magrittr.
Ooms, Jeroen. 2014. “The Jsonlite Package: A Practical and Consistent Mapping Between Json Data and R Objects.” arXiv:1403.2805 [stat.CO]. http://arxiv.org/abs/1403.2805.
Vaidyanathan, Ramnath, Yihui Xie, JJ Allaire, Joe Cheng, and Kenton Russell. 2016. Htmlwidgets: HTML Widgets for R. https://CRAN.R-project.org/package=htmlwidgets.
Pedersen, Thomas Lin. 2019. Ggforce: Accelerating ’Ggplot2’. https://CRAN.R-project.org/package=ggforce.
Tierney, Nicholas, Di Cook, Miles McBain, and Colin Fay. 2018. Naniar: Data Structures, Summaries, and Visualisations for Missing Data. https://github.com/njtierney/naniar.
Schloerke, Barret, Jason Crowley, Di Cook, Francois Briatte, Moritz Marbach, Edwin Thoen, Amos Elberg, and Joseph Larmarange. 2016. GGally: Extension to ’Ggplot2’.
Harrell Jr, Frank E, with contributions from Charles Dupont, and many others. 2019. Hmisc: Harrell Miscellaneous. https://CRAN.R-project.org/package=Hmisc.
Wickham, Hadley, and Garrett Grolemund. 2018. R for Data Science. O’Reilly.
If you aren’t already familiar with the grammar of graphics or ggplot2, we recommend reading the Data Visualization chapter from the R for Data Science book. https://r4ds.had.co.nz/data-visualisation.html↩
Technically speaking, these dplyr verbs are S3 generic functions that plotly has a defined a custom method for. In nearly every case, that custom method simply queries the data underlying the plotly object, applies the dplyr function, then adds the transformed data back into the resulting plotly object↩
A trace is similar in concept to a layer (as defined in Section 2.1, but it’s not quite the same. In many cases, like the bottom panel of Figure 2.1, it makes sense to implement a single layer as multiple traces. This is due to the design of plotly.js and how traces are tied to legends and hover behavior.↩
The htmlwidgets package provides a foundation for other packages to implement R bindings to JavaScript libraries so that those bindings work in various contexts (e.g. the R console, RStudio, inside rmarkdown documents, shiny apps, etc). For more info and examples, see the website http://www.htmlwidgets.org.↩
Although the x-axis is discrete, plotly.js still considers this a histogram because it’s generating it generates counts in the browser. Learn more about the difference between histograms and bar charts in section 5.↩
This section is not meant to teach you ggplot2, but rather to help point out when and why it might be preferrable to
plot_ly(). If you’re new to ggplot2 and would like to learn it, see 1.3.3.↩As discussed in 34,
ggplotly()can actually convert custom geoms as well, but each one requires a custom hook, and many custom geoms are not yet supported.↩